-Search query
-Search result
Showing 1 - 50 of 82 items for (author: liu & js)

EMDB-41140: 
APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD
Method: single particle / : Bodrug T, Welsh KA, Bolhuis DL, Paulakonis E, Martinez-Chacin RC, Liu B, Pinkin N, Bonacci T, Cui L, Xu P, Roscow O, Amann SJ, Grishkovskaya I, Emanuele MJ, Harrison JS, Steimel JP, Hahn KM, Zhang W, Zhong E, Haselbach D, Brown NG

EMDB-41142: 
APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB
Method: single particle / : Bodrug T, Welsh KA, Bolhuis DL, Paulakonis E, Martinez-Chacin RC, Liu B, Pinkin N, Bonacci T, Cui L, Xu P, Roscow O, Amann SJ, Grishkovskaya I, Emanuele MJ, Harrison JS, Steimel JP, Hahn KM, Zhang W, Zhong E, Haselbach D, Brown NG

EMDB-16144: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the macrocyclic peptide S1B3inL1
Method: single particle / : Hurdiss DL, Drulyte I

PDB-8bon: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the macrocyclic peptide S1B3inL1
Method: single particle / : Hurdiss DL

EMDB-28178: 
Structure of lineage IV Lassa virus glycoprotein complex (strain Josiah)
Method: single particle / : Perrett HR, Ward AB

EMDB-28179: 
Structure of lineage II Lassa virus glycoprotein complex (strain NIG08-A41)
Method: single particle / : Perrett HR, Ward AB

EMDB-28180: 
Structure of lineage V Lassa virus glycoprotein complex (strain Soromba-R)
Method: single particle / : Perrett HR, Ward AB

EMDB-28181: 
Structure of lineage VII Lassa virus glycoprotein complex (strain Togo/2016/7082)
Method: single particle / : Perrett HR, Ward AB

EMDB-28182: 
Lassa virus glycoprotein complex (Josiah) bound to 12.1F Fab
Method: single particle / : Perrett HR, Ward AB

EMDB-28183: 
Lassa virus glycoprotein complex (Josiah) bound to 19.7E Fab
Method: single particle / : Perrett HR, Ward AB

EMDB-28184: 
Lassa virus glycoprotein complex (Josiah) bound to S370.7 Fab
Method: single particle / : Perrett HR, Ward AB

PDB-8ejd: 
Structure of lineage IV Lassa virus glycoprotein complex (strain Josiah)
Method: single particle / : Perrett HR, Ward AB

PDB-8eje: 
Structure of lineage II Lassa virus glycoprotein complex (strain NIG08-A41)
Method: single particle / : Perrett HR, Ward AB

PDB-8ejf: 
Structure of lineage V Lassa virus glycoprotein complex (strain Soromba-R)
Method: single particle / : Perrett HR, Ward AB

PDB-8ejg: 
Structure of lineage VII Lassa virus glycoprotein complex (strain Togo/2016/7082)
Method: single particle / : Perrett HR, Ward AB

PDB-8ejh: 
Lassa virus glycoprotein complex (Josiah) bound to 12.1F Fab
Method: single particle / : Perrett HR, Ward AB

PDB-8eji: 
Lassa virus glycoprotein complex (Josiah) bound to 19.7E Fab
Method: single particle / : Perrett HR, Ward AB

PDB-8ejj: 
Lassa virus glycoprotein complex (Josiah) bound to S370.7 Fab
Method: single particle / : Perrett HR, Ward AB

EMDB-17154: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (consensus and constituent map 1)
Method: single particle / : Stoos L, Michael AK, Kempf G, Cavadini S, Thoma NH

EMDB-17155: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1)
Method: single particle / : Michael AK, Stoos L, Kempf G, Cavadini S, Thoma NH

EMDB-17156: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 2)
Method: single particle / : Michael AK, Stoos L, Kempf G, Cavadini S, Thoma NH

EMDB-17157: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map)
Method: single particle / : Stoos L, Michael AK, Kempf G, Cavadini S, Thoma NH

EMDB-17158: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (constituent map 2 from additional focus classification on PAS domains)
Method: single particle / : Stoos L, Michael AK, Kempf G, Cavadini S, Thoma NH

EMDB-17159: 
Cryo-EM map of MYC-MAX-OCT4-LIN28 complex
Method: single particle / : Michael AK, Kempf G, Cavadini S, Thoma NH

EMDB-17160: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement)
Method: single particle / : Michael AK, Stoos L, Kempf G, Cavadini S, Thoma N

EMDB-17161: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 1)
Method: single particle / : Michael AK, Stoos L, Cavadini S, Kempf G

EMDB-17162: 
MAX-MAX bound to a nucleosome at SHL+5.1 and SHL-6.9.
Method: single particle / : Stoos L, Kempf G, Kater L, Thoma NH

EMDB-17183: 
OCT4 and MYC-MAX co-bound to a nucleosome
Method: single particle / : Michael AK, Stoos L, Kempf G, Cavadini S, Thoma N

EMDB-17184: 
MYC-MAX bound to a nucleosome at SHL+5.8
Method: single particle / : Stoos L, Michael AK, Kempf G, Kater L, Cavadini S, Thoma N

EMDB-28669: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 01)
Method: electron tomography / : Liu J, Ren G
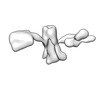
EMDB-28670: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 02)
Method: electron tomography / : Liu J, Ren G
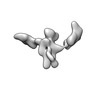
EMDB-28671: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 03)
Method: electron tomography / : Liu J, Ren G
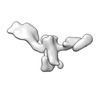
EMDB-28672: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 04)
Method: electron tomography / : Liu J, Ren G

EMDB-28673: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 05)
Method: electron tomography / : Liu J, Ren G
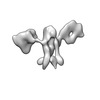
EMDB-28674: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 06)
Method: electron tomography / : Liu J, Ren G
EMDB-28675: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 07)
Method: electron tomography / : Liu J, Ren G
EMDB-28676: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 08)
Method: electron tomography / : Liu J, Ren G

EMDB-28677: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 09)
Method: electron tomography / : Liu J, Ren G

EMDB-28678: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 10)
Method: electron tomography / : Liu J, Ren G

EMDB-28679: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 11)
Method: electron tomography / : Liu J, Ren G

EMDB-28680: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 12)
Method: electron tomography / : Liu J, Ren G

EMDB-28681: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 13)
Method: electron tomography / : Liu J, Ren G

EMDB-28682: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 14)
Method: electron tomography / : Liu J, Ren G
EMDB-28683: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 15)
Method: electron tomography / : Liu J, Ren G
EMDB-28684: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 16)
Method: electron tomography / : Liu J, Ren G

EMDB-16511: 
HERV-K Gag immature lattice
Method: subtomogram averaging / : Krebs AS, Liu HF, Zhou Y, Rey JS, Levintov L, Perilla JR, Bartesaghi A, Zhang P

EMDB-13926: 
Twist-corrected RNA origami 5-helix Tile A
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13625: 
Mature conformer 2 of a 6-helix bundle of RNA with a clasp
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13626: 
Young conformer 2 of a 6-helix bundle of RNA with a clasp
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13627: 
6-Helix bundle of RNA
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
